
Headlines like this have become all but common within the healthcare space for the last 1-2 years.
The fear is real. I understand it. So let’s delve into what’s happening.
First, what are LLMs?
Large Language Models (LLMs) are Artificial Intelligence systems that have learned to systematize and write human language by ingesting huge amounts of text—like books, articles, and websites. Think of them like very smart “text predictors” that use statistics to guess the next word in a sentence based on what came before. By practicing this guessing game many billions of times, they get really good at creating sentences that make sense.
These models use special section of Artificial Intelligence called transformers that helps them understand context – how words connect to each other in long sentences or paragraphs. This means they can answer questions, summarize information, or even write new text that sounds natural.
In healthcare and computational biology, LLMs can be helpful to scientists and doctors by quickly reading and summarizing tons of medical papers or patient records, assisting in writing reports and other tedious tasks…
They do not “think” like people, they are essentially pattern seeking machines that use patterns they learned during training to generate useful language. Sometimes their answers can surprise people by being very creative or very wrong, so humans need to ALWAYS check their work.
But can they be useful? We’re gonna explore some usages of LLMs in medicine and some benefits and drawbacks from that use.
Usages of LLMs in Healthcare
- Clinical Decision Support: LLMs like GPT-3.5 and GPT-4 analyze electronic health records and medical imaging results to assist clinicians with diagnosis and treatment planning. For example, LLMs have been used as medical chatbots across many specialties, helping interpret patient data and medical literature. (Source)
- Medical Education and Research: LLMs facilitate rapid literature review, summarize complex biomedical information, and aid in clinical trial design and patient recruitment, supporting research workflows in computational biology and medicine. (Source)
- Patient Communication: LLMs generate personalized, understandable explanations for patients, convert complex medical jargon into plain language, and provide tailored health guidance to empower patients and caregivers. (Source)
- Health Promotion and Disease Prevention: By integrating patient data, LLMs identify high-risk groups and recommend preventive strategies, fostering proactive medical care. (Source)
- Clinical Workflow Automation: Tasks such as summarizing discharge instructions and generating clinical documentation have been streamlined with LLM support. (Source)
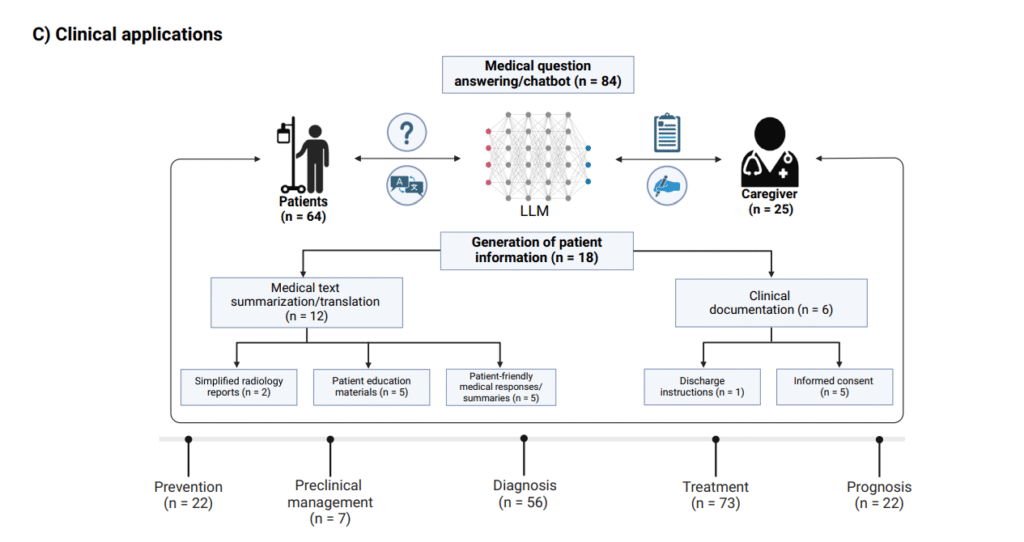
Let’s talk first about the benefits, I understand the fears, but to properly address the usage, we also need to understand the benefits:
- Efficiency and Scalability: LLMs can process massive datasets quickly, enabling faster clinical decision-making and literature synthesis.
- Personalization: Models will be able in the future to provide individualized patient advice based on medical history and current guidelines, enhancing treatment adherence.
- Broadened Access: LLM-driven chatbots can become educational tools to increase healthcare accessibility, especially in remote or underserved areas.
- Advanced Research Tools: They are a massive support in Literature and Database tasks such as drug discovery and gene function prediction.
Finally, let’s discuss the dangers:
- Diagnostic Limitations: LLMs can have severe limitations and are prone to hallucinations- Despite successes on exams like the USMLE, LLMs can lack clinical context and reasoning, leading to errors in real-world settings.
- Lack of Transparency: Proprietary models often do not disclose training data, impeding trust and validation by clinicians. This is why public research is paramount and so important to counteract proprietary models;
- Non-reproducibility: Model output may vary across runs due to inherent stochasticity, which presents clinical risk, especially for minority groups which are underepressented in the data;
- Bias and Safety Concerns: Training data bias can lead to inaccurate or harmful recommendations; hallucinations (fabricated content) remain a frequent issue. Minorities within the data tend to see the biases they face reflected on the LLM.
- Technical and Ethical Issues: Data privacy, the need for human oversight, and evolving regulatory frameworks pose challenges for widespread adoption.
LLMs represent a transformative, powerful tool that can augment healthcare professionals and researchers, but their effective and safe use depends on awareness of their strengths and limitations. Human expertise remains essential to interpret, verify, and ethically integrate these AI systems at the frontlines of medicine.




